Calculating the “rank score” tracks
Our browser provides two tracks for each neutrality test, one of the “scores” and one for the “ranked scores”. The purpose of this latter type of tracks, the ranked scores, is to help identify which SNPs or regions have significantly high scores compared to the rest of genome. So, if you are interested in knowing if if any of the SNPs in your favorite gene have a significantly high score, the best way to do it is by looking at the “ranked scores” tracks.
Our Rank Scores are genome based rank “p-values“, meaning that we calculate them based on the rank of a score in the genomic distribution. So, if a region has a large rank score, it means that its score belongs to the extreme upper (or lower, depending on the test) tail of the genome-wide distribution. The top score in the genome has a rank-score of almost 0 (actually 1/N; with N the number of scores in the genome-wide distribution); and the lowest score has a p-value of 1.
The method
The following chart describes our workflow to calculate rank scores:
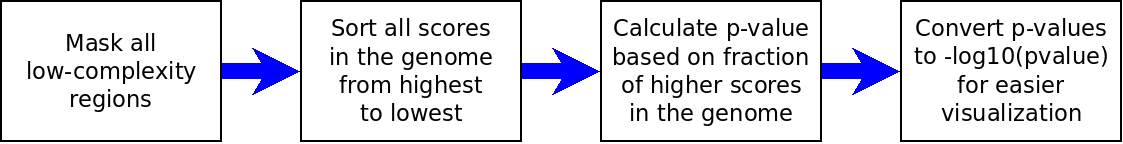 Workflow to calculate p-values in our pipeline.
Workflow to calculate p-values in our pipeline.
The first step is to remove all the low complexity regions, such as centromers and telomers. In some cases, we also remove regions like the HLA region, because they are difficult to annotate. This masking step is needed because these regions tend to have aberrant scores, and may produce false positives, which will invalidate the calculation of rank scores.
The second step is simply to sort all the scores, from the highest to the lowest. This way, we get a sorted list of all the scores in a distribution, for a given population. The only exceptions are the cases of Tajima’s D, Fay & Wu’s H, Fu’s F, Fu and Li’s F*, Fu and Li’s D*, Dh, R2 and pi, for which we sorted the scores from the lowest to the highest. This is because for these tests, low scores are expected under positive selection.
The third step is to calculate the rank scores. In this case, the rank score is just the fraction of values that are higher than the given score in the genomic distribution. For example, the region that has the top score in the genome has a p-value of almost 0, because no other region has a score higher than it. As another example, if a region has a score that is the 50th score of the distribution, and there are 1000 regions in our list, that region will have a rank score of 0.05. As a last example, the region that has the lowest score in the genome will have a p-value of 1.
 Example of how p-values are calculated in our pipeline. In this case, we assume that there are only 200 SNPs in the genome. Note that SNPs having the same score (3rd and 4th rows) have also the same p-value.
Example of how p-values are calculated in our pipeline. In this case, we assume that there are only 200 SNPs in the genome. Note that SNPs having the same score (3rd and 4th rows) have also the same p-value.
The last step is to convert all the rank scores to -log10(rank score). This is done in order to improve the visualization of scores in the browser tracks. Thanks to this transformation, low p-values become large values, so it is easier to spot them in the tracks.
Note that after the transformation of rank score to log scores, a rank score of 0.01 becomes equal to 2. So, if you want to use the Genome Browser to determine which regions have a significant rank score (at 0.01 false discovery rate) according to the rest of the genome, you should look for p-values higher than 2.
Pros and Cons of the method
A good discussion on the advantages and drawbacks of our approach is in the paper Teshima et al. 2009 . Here, we will describe a couple of drawbacks.
One disadvantage of this method is that it is sensitive to false positives and artifacts. If for some reason one test for selection gives high values for regions of low complexity, these scores, which are artifacts, will reduce the rank score of all the other scores in the genome. For this reason we did a masking of all the low-complexity regions before calculating the rank scores. However, we may have missed some regions.
Another disadvantage of this method is that it only takes into account the rank of a score in the distribution, and not its value. So, if the top score in the genome is 1000, and the second is 100, the p-value of the second will still be 0.02, regardless of the fact that the difference between these two scores is huge. Nevertheless, this problem should affect only a few scores at the top of the distribution, and it can be avoided by looking at the corresponding scores track in the browser.
Last, this method requires that you fix a priori a percentage of the genome you expect to have evolve under positive selection. Indeed, if you fix a threshold of 5% for your rank score, you will obtain exactly 5% of the genome to be the target of positive selection with this criteria.
Other approaches to calculate rank scores
In the scientific literature, there are other ways to calculate rank scores. In our case, we had to choose a method that could be applied easily to all the tests, and that could be understood by everyone. However, let’s describe a few alternatives to rank scores.
One method is to compare the scores against simulated data. We can use a software like cosi to simulate thousands of populations having the same demographic history as the populations in 1000genomes. Then, we can calculate the rank scores by calculating the rank of each score, but using the distribution of values in all the simulations instead of using only the genome. The major drawback of this method is that is sensible to the demographic model used to make the simulations: so, depending on our hypothesis on how human populations have migrated, we can get different p-values. We will provide rank scores based on simulations in a future release of our data, but for the first release, we preferred to provide values that are not dependent on any assumption on population history.
Another method, is to calculate formal p-values by assuming that the scores have a given distribution, like a normal or a exponential distribution. For example, iHS scores are, by definition, distributed normally. So, to calculate the p-value of a iHS score, we may calculate the probability of observing that score in a normal distribution. The drawback of this method is that most of the tests we implemented are not distributed normally. So, to apply this method, we would have to find a way to normalize the distribution of each test. This is a risky step, because there are usually many alternative approaches to normalize a dataset, and if we make the wrong decision we may end up having wrong p-values. So, for the first release of the dataset, we decided not to take the risk of transforming our data, and we preferred using the method of ranked p-values.