Here is a list of the tests implemented in our pipeline.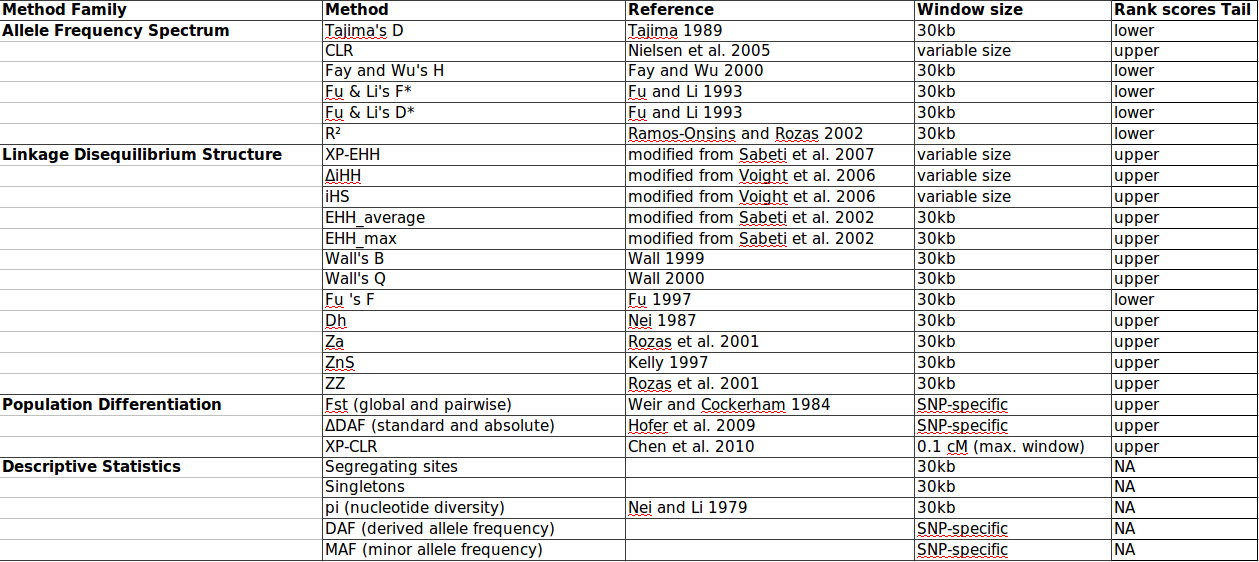
- Tajima’s D (Tajima, 1989): Comparison of estimates of the number of segregating sites and the mean pairwise difference between sequences.
- CLR (Nielsen et al., 2005). Multilocus Composite Likelihood Ratio Test. Read more about the CLR
- Fay and Wu’s H (Fay & Wu, 2000): Comparison of the number of derived segregating sites at low and high frequencies and the number of variants at intermediate frequencies.
- Fu and Li’s F* (Fu, 1997): Comparison of the number of singleton mutations and the mean pairwise difference between sequences.
- Fu and Li’s D* (Fu, 1997): Comparison of the number of singleton mutations and the total number of nucleotide variants.
- R2 (Ramos-Onsins and Rozas. 2002) Comparison of the difference between the number of singletons per sequence and the average number of nucleotide differences.
- XP-EHH (Sabeti et al., 2007): Cross-population extended haplotype homozygosity.
- Delta iHH (Voight et al., 2006, Grossman et al., 2010): difference between two integrated haplotype homozygosity scores.
- iHS (Voight et al., 2006): log ratio between two integrated haplotype homozygosity scores.
- EHH average (Sabeti et al., 2002): Extended halotype homozygosity; weighted average for all core haplotypes of the position at which the haplotype homozygosity decays to <=0.5.
- Wall’s B (Wall, 2000): Counts the number of pairs of adjacent segregating sites that are congruent (if the subset of the data consisting of the two sites contains only two different haplotypes)
- Wall’s Q (Wall, 2000): Adds the number of partitions (two disjoint subsets whose union is the set of individuals in the sample) induced by congruent pairs to Wall’s B.
- Fu’s Fs (Fu, 1997): Based on Ewens’ sampling distribution, taking into account the number of different haplotypes in the sample.
- Dh (Nei, 1987): Summary statistic based on the number of different haplotypes in the sample
- Fst (Weir and Cockerham, 1984) : global and pairwise
- delta DAF: difference of Derived allele frequencies between 2 populations.
- XP-CLR (Chen et al., 2010): Multilocus allele frequency differentiation between two populations.